Note
Go to the end to download the full example code or to run this example in your browser via Binder
Scikit-learn hyperparameter search wrapper#
Iaroslav Shcherbatyi, Tim Head and Gilles Louppe. June 2017. Reformatted by Holger Nahrstaedt 2020
Introduction#
This example assumes basic familiarity with scikit-learn.
Search for parameters of machine learning models that results in best
cross-validation performance is necessary in almost all practical
cases to get a model with best generalization estimate.
A standard approach in scikit-learn is to use
sklearn.model_selection.GridSearchCV class, which enumerates
all combinations of hyperparameters values given as input.
This search complexity grows exponentially with the number of parameters.
A more scalable approach is to use
sklearn.model_selection.RandomizedSearchCV, which however does not
take advantage of the structure of a search space.
Scikit-optimize provides a drop-in replacement for these two scikit-learn
methods. The hyperparameter search is achieved by Bayesian Optimization
At each step of the optimization, a surrogate model infers the objective
function using observed evluation results as priors. An acquisition function
utilizes these predictions to navigate between exploration (sampling
unexplored areas) and exploitation (focusing on regions likely containing
the global optimum). By balancing these two strategies, Bayesian Optimization
identifies probable optimal areas while ensuring comprehensive search
coverage.
In practice, this method often leads to quicker and better results.
Note: for a manual hyperparameter optimization example, see “Hyperparameter Optimization” notebook.
print(__doc__)
import numpy as np
np.random.seed(123)
import matplotlib.pyplot as plt
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
from skopt import BayesSearchCV
Minimal example#
A minimal example of optimizing hyperparameters of SVC (Support Vector machine Classifier) is given below.
X, y = load_digits(n_class=10, return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.75, test_size=0.25, random_state=0
)
# log-uniform: understand as search over p = exp(x) by varying x
opt = BayesSearchCV(
SVC(),
{
'C': (1e-6, 1e6, 'log-uniform'),
'gamma': (1e-6, 1e1, 'log-uniform'),
'degree': (1, 8), # integer valued parameter
'kernel': ['linear', 'poly', 'rbf'], # categorical parameter
},
n_iter=32,
cv=3,
)
opt.fit(X_train, y_train)
print("val. score: %s" % opt.best_score_)
print("test score: %s" % opt.score(X_test, y_test))
val. score: 0.9866369710467705
test score: 0.9844444444444445
Advanced example#
In practice, one wants to enumerate over multiple predictive model classes, with different search spaces and number of evaluations per class. An example of such search over parameters of Linear SVM, Kernel SVM, and decision trees is given below.
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.svm import SVC, LinearSVC
from skopt import BayesSearchCV
from skopt.plots import plot_histogram, plot_objective
from skopt.space import Categorical, Integer, Real
X, y = load_digits(n_class=10, return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
# pipeline class is used as estimator to enable
# search over different model types
pipe = Pipeline([('model', SVC())])
# single categorical value of 'model' parameter is
# sets the model class
# We will get ConvergenceWarnings because the problem is not well-conditioned.
# But that's fine, this is just an example.
linsvc_search = {
'model': [LinearSVC(max_iter=1000, dual="auto")],
'model__C': (1e-6, 1e6, 'log-uniform'),
}
# explicit dimension classes can be specified like this
svc_search = {
'model': Categorical([SVC()]),
'model__C': Real(1e-6, 1e6, prior='log-uniform'),
'model__gamma': Real(1e-6, 1e1, prior='log-uniform'),
'model__degree': Integer(1, 8),
'model__kernel': Categorical(['linear', 'poly', 'rbf']),
}
opt = BayesSearchCV(
pipe,
# (parameter space, # of evaluations)
[(svc_search, 40), (linsvc_search, 16)],
cv=3,
)
opt.fit(X_train, y_train)
print("val. score: %s" % opt.best_score_)
print("test score: %s" % opt.score(X_test, y_test))
print("best params: %s" % str(opt.best_params_))
val. score: 0.9881217520415739
test score: 0.9888888888888889
best params: OrderedDict([('model', SVC()), ('model__C', 4.580543393203649), ('model__degree', 3), ('model__gamma', 0.0002585937465230229), ('model__kernel', 'poly')])
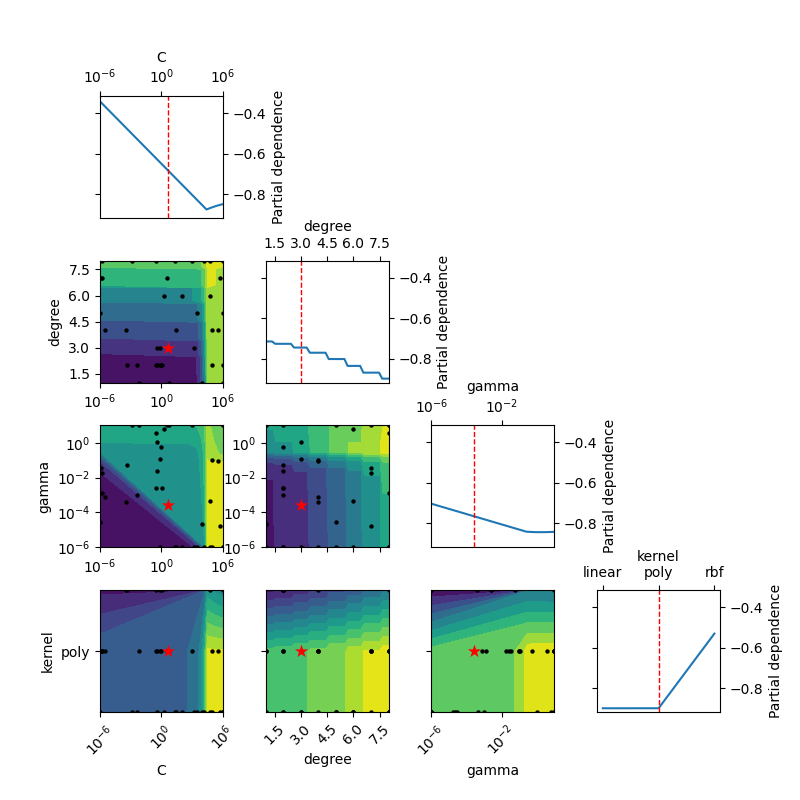
Partial Dependence plot of the objective function for SVC
_ = plot_objective(
opt.optimizer_results_[0],
dimensions=["C", "degree", "gamma", "kernel"],
n_minimum_search=int(1e8),
)
plt.show()
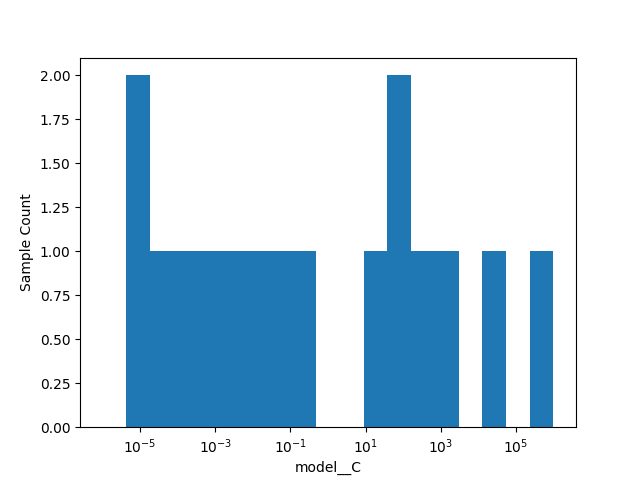
Plot of the histogram for LinearSVC
_ = plot_histogram(opt.optimizer_results_[1], 1)
plt.show()
Progress monitoring and control using callback argument of fit method#
It is possible to monitor the progress of BayesSearchCV with an event
handler that is called on every step of subspace exploration. For single job
mode, this is called on every evaluation of model configuration, and for
parallel mode, this is called when n_jobs model configurations are evaluated
in parallel.
Additionally, exploration can be stopped if the callback returns True.
This can be used to stop the exploration early, for instance when the
accuracy that you get is sufficiently high.
An example usage is shown below.
from sklearn.datasets import load_iris
from sklearn.svm import SVC
from skopt import BayesSearchCV
X, y = load_iris(return_X_y=True)
searchcv = BayesSearchCV(
SVC(gamma='scale'),
search_spaces={'C': (0.01, 100.0, 'log-uniform')},
n_iter=10,
cv=3,
)
# callback handler
def on_step(optim_result):
score = -optim_result['fun']
print("best score: %s" % score)
if score >= 0.98:
print('Interrupting!')
return True
searchcv.fit(X, y, callback=on_step)
best score: 0.98
Interrupting!
Counting total iterations that will be used to explore all subspaces#
Subspaces in previous examples can further increase in complexity if you add
new model subspaces or dimensions for feature extraction pipelines. For
monitoring of progress, you would like to know the total number of
iterations it will take to explore all subspaces. This can be
calculated with total_iterations property, as in the code below.
from sklearn.datasets import load_iris
from sklearn.svm import SVC
from skopt import BayesSearchCV
X, y = load_iris(return_X_y=True)
searchcv = BayesSearchCV(
SVC(),
search_spaces=[
({'C': (0.1, 1.0)}, 19), # 19 iterations for this subspace
{'gamma': (0.1, 1.0)},
],
n_iter=10,
)
print(searchcv.total_iterations)
29
Total running time of the script: (1 minutes 33.880 seconds)
